Indexing, Slicing and Subsetting DataFrames in Python
Last updated on 2024-02-26 | Edit this page
Estimated time: 60 minutes
Overview
Questions
- How can I access specific data within my data set?
- How can Python and Pandas help me to analyse my data?
Objectives
- Describe what 0-based indexing is.
- Manipulate and extract data using column headings and index locations.
- Employ slicing to select sets of data from a DataFrame.
- Employ label and integer-based indexing to select ranges of data in a dataframe.
- Reassign values within subsets of a DataFrame.
- Create a copy of a DataFrame.
- Query / select a subset of data using a set of criteria using the
following operators:
==,!=,>,<,>=,<=. - Locate subsets of data using masks.
- Describe BOOLEAN objects in Python and manipulate data using BOOLEANs.
Tip: use .head() method throughout this lesson to keep
your display neater for students. Encourage students to try with and
without .head() to reinforce this useful tool and then to
use it or not at their preference.
For example, if a student worries about keeping up in pace with
typing, let them know they can skip the .head(), but that
you’ll use it to keep more lines of previous steps visible.
In the first episode of this lesson, we read a CSV file into a pandas’ DataFrame. We learned how to:
- save a DataFrame to a named object,
- perform basic math on data,
- calculate summary statistics, and
- create plots based on the data we loaded into pandas.
In this lesson, we will explore ways to access different parts of the data using:
- indexing,
- slicing, and
- subsetting.
Loading our data
We will continue to use the surveys dataset that we worked with in the last episode. Let’s reopen and read in the data again:
Indexing and Slicing in Python
We often want to work with subsets of a DataFrame object. There are different ways to accomplish this including: using labels (column headings), numeric ranges, or specific x,y index locations.
Selecting data using Labels (Column Headings)
We use square brackets [] to select a subset of a Python
object. For example, we can select all data from a column named
species_id from the surveys_df DataFrame by
name. There are two ways to do this:
PYTHON
# TIP: use the .head() method we saw earlier to make output shorter
# Method 1: select a 'subset' of the data using the column name
surveys_df['species_id']
# Method 2: use the column name as an 'attribute'; gives the same output
surveys_df.species_idWe can also create a new object that contains only the data within
the species_id column as follows:
PYTHON
# Creates an object, surveys_species, that only contains the `species_id` column
surveys_species = surveys_df['species_id']We can pass a list of column names too, as an index to select columns in that order. This is useful when we need to reorganize our data.
NOTE: If a column name is not contained in the DataFrame, an exception (error) will be raised.
PYTHON
# Select the species and plot columns from the DataFrame
surveys_df[['species_id', 'plot_id']]
# What happens when you flip the order?
surveys_df[['plot_id', 'species_id']]
# What happens if you ask for a column that doesn't exist?
surveys_df['speciess']Python tells us what type of error it is in the traceback, at the
bottom it says KeyError: 'speciess' which means that
speciess is not a valid column name (nor a valid key in the
related Python data type dictionary).
Reminder
The Python language and its modules (such as Pandas) define reserved
words that should not be used as identifiers when assigning objects and
variable names. Examples of reserved words in Python include Boolean
values True and False, operators
and, or, and not, among others.
The full list of reserved words for Python version 3 is provided at https://docs.python.org/3/reference/lexical_analysis.html#identifiers.
When naming objects and variables, it’s also important to avoid using
the names of built-in data structures and methods. For example, a
list is a built-in data type. It is possible to use the word
‘list’ as an identifier for a new object, for example
list = ['apples', 'oranges', 'bananas']. However, you would
then be unable to create an empty list using list() or
convert a tuple to a list using list(sometuple).
Extracting Range based Subsets: Slicing
Reminder
Python uses 0-based indexing.
Let’s remind ourselves that Python uses 0-based indexing. This means
that the first element in an object is located at position
0. This is different from other tools like R and Matlab
that index elements within objects starting at 1.
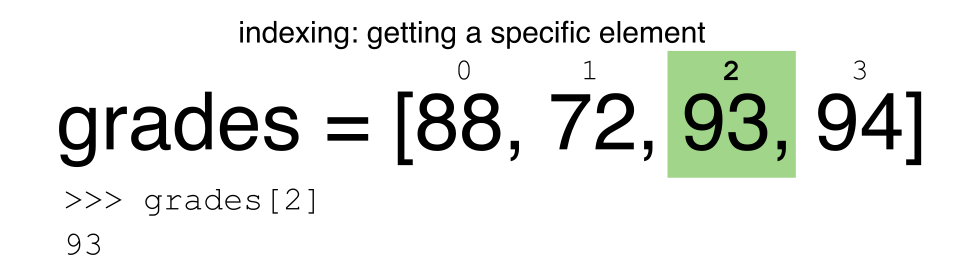
a[0]returns1, as Python starts with element 0 (this may be different from what you have previously experience with other languages e.g. MATLAB and R)a[5]raises anIndexErrorThe error is raised because the list
ahas no element with index 5: it has only five entries, indexed from 0 to 4.-
a[len(a)]also raises anIndexError.len(a)returns5, makinga[len(a)]equivalent toa[5]. To retreive the final element of a list, us the index-1, e.g.OUTPUT
5
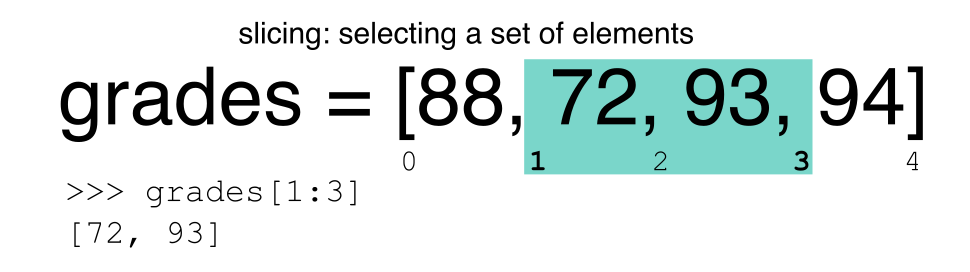
Slicing Subsets of Rows in Python
Slicing using the [] operator selects a set of rows
and/or columns from a DataFrame. To slice out a set of rows, you use the
following syntax: data[start:stop]. When slicing in pandas
the start bound is included in the output. The stop bound is one step
BEYOND the row you want to select. So if you want to select rows 0, 1
and 2 your code would look like this:
The stop bound in Python is different from what you might be used to in languages like Matlab and R.
PYTHON
# Select the first 5 rows (rows 0, 1, 2, 3, 4)
surveys_df[:5]
# Select the last element in the list
# (the slice starts at the last element, and ends at the end of the list)
surveys_df[-1:]We can also reassign values within subsets of our DataFrame.
But before we do that, let’s look at the difference between the concept of copying objects and the concept of referencing objects in Python.
Copying Objects vs Referencing Objects in Python
Let’s start with an example:
PYTHON
# Using the 'copy() method'
true_copy_surveys_df = surveys_df.copy()
# Using the '=' operator
ref_surveys_df = surveys_dfYou might think that the code
ref_surveys_df = surveys_df creates a fresh distinct copy
of the surveys_df DataFrame object. However, using the
= operator in the simple statement y = x does
not create a copy of our DataFrame. Instead,
y = x creates a new variable y that references
the same object that x refers to. To state
this another way, there is only one object (the
DataFrame), and both x and y refer to it.
In contrast, the copy() method for a DataFrame creates a
true copy of the DataFrame.
Let’s look at what happens when we reassign the values within a subset of the DataFrame that references another DataFrame object:
PYTHON
# Assign the value `0` to the first three rows of data in the DataFrame
ref_surveys_df[0:3] = 0Let’s try the following code:
PYTHON
# ref_surveys_df was created using the '=' operator
ref_surveys_df.head()
# true_copy_surveys_df was created using the copy() function
true_copy_surveys_df.head()
# surveys_df is the original dataframe
surveys_df.head()What is the difference between these three dataframes?
When we assigned the first 3 rows the value of 0 using
the ref_surveys_df DataFrame, the surveys_df
DataFrame is modified too. Remember we created the reference
ref_surveys_df object above when we did
ref_surveys_df = surveys_df. Remember
surveys_df and ref_surveys_df refer to the
same exact DataFrame object. If either one changes the object, the other
will see the same changes to the reference object.
However - true_copy_surveys_df was created via the
copy() function. It retains the original values for the
first three rows.
To review and recap:
-
Copy uses the dataframe’s
copy()method -
A Reference is created using the
=operator
Okay, that’s enough of that. Let’s create a brand new clean dataframe from the original data CSV file.
Slicing Subsets of Rows and Columns in Python
We can select specific ranges of our data in both the row and column directions using either label or integer-based indexing.
ilocis primarily an integer based indexing counting from 0. That is, your specify rows and columns giving a number. Thus, the first row is row 0, the second column is column 1, etc.locis primarily a label based indexing where you can refer to rows and columns by their name. E.g., column ‘month’. Note that integers may be used, but they are interpreted as a label.
To select a subset of rows and columns from our
DataFrame, we can use the iloc method. For example, we can
select month, day and year (columns 2, 3 and 4 if we start counting at
1), like this:
which gives the output
OUTPUT
month day year
0 7 16 1977
1 7 16 1977
2 7 16 1977Notice that we asked for a slice from 0:3. This yielded 3 rows of data. When you ask for 0:3, you are actually telling Python to start at index 0 and select rows 0, 1, 2 up to but not including 3.
Let’s explore some other ways to index and select subsets of data:
PYTHON
# Select all columns for rows of index values 0 and 10
surveys_df.loc[[0, 10], :]
# What does this do?
surveys_df.loc[0, ['species_id', 'plot_id', 'weight']]
# What happens when you type the code below?
surveys_df.loc[[0, 10, 35549], :]NOTE: Labels must be found in the DataFrame or you
will get a KeyError.
Indexing by labels loc differs from indexing by integers
iloc. With loc, both the start bound and the
stop bound are inclusive. When using loc,
integers can be used, but the integers refer to the index label
and not the position. For example, using loc and select 1:4
will get a different result than using iloc to select rows
1:4.
We can also select a specific data value using a row and column
location within the DataFrame and iloc indexing:
In this iloc example,
gives the output
OUTPUT
'F'Remember that Python indexing begins at 0. So, the index location [2, 6] selects the element that is 3 rows down and 7 columns over in the DataFrame.
It is worth noting that rows are selected when using loc
with a single list of labels (or iloc with a single list of
integers). However, unlike loc or iloc,
indexing a data frame directly with labels will select columns (e.g.
surveys_df[['species_id', 'plot_id', 'weight']]), while
ranges of integers will select rows
(e.g. surveys\_df[0:13]). Direct indexing of rows is
redundant with using iloc, and will raise a
KeyError if a single integer or list is used; the error
will also occur if index labels are used without loc (or
column labels used with it). A useful rule of thumb is the following:
integer-based slicing is best done with iloc and will avoid
errors (and is generally consistent with indexing of Numpy arrays),
label-based slicing of rows is done with loc, and slicing
of columns by directly indexing column names.
Challenge - Range
- What happens when you execute:
surveys_df[0:1]surveys_df[0]surveys_df[:4]surveys_df[:-1]
- What happens when you call:
surveys_df.iloc[0:1]surveys_df.iloc[0]surveys_df.iloc[:4, :]surveys_df.iloc[0:4, 1:4]surveys_df.loc[0:4, 1:4]
- How are the last two commands different?
-
-
surveys_df[0:3]returns the first three rows of the DataFrame:
OUTPUT
record_id month day year plot_id species_id sex hindfoot_length weight 0 1 7 16 1977 2 NL M 32.0 NaN 1 2 7 16 1977 3 NL M 33.0 NaN 2 3 7 16 1977 2 DM F 37.0 NaN-
surveys_df[0]results in a ‘KeyError’, since direct indexing of a row is redundant withiloc. -
surveys_df[0:1]can be used to obtain only the first row. -
surveys_df[:5]slices from the first row to the fifth:
OUTPUT
record_id month day year plot_id species_id sex hindfoot_length weight 0 1 7 16 1977 2 NL M 32.0 NaN 1 2 7 16 1977 3 NL M 33.0 NaN 2 3 7 16 1977 2 DM F 37.0 NaN 3 4 7 16 1977 7 DM M 36.0 NaN 4 5 7 16 1977 3 DM M 35.0 NaN-
surveys_df[:-1]provides everything except the final row of the DataFrame. You can use negative index numbers to count backwards from the last entry.
-
-
surveys_df.iloc[0:1]returns the first row -
surveys_df.iloc[0]returns the first row as a named list -
surveys_df.iloc[:4, :]returns all columns of the first four rows -
surveys_df.iloc[0:4, 1:4]selects specified columns of the first four rows -
surveys_df.loc[0:4, 1:4]results in a ‘TypeError’ - see below.
-
- While
ilocuses integers as indices and slices accordingly,locworks with labels. It is like accessing values from a dictionary, asking for the key names. Column names 1:4 do not exist, so the call tolocabove results in an error. Check also the difference betweensurveys_df.loc[0:4]andsurveys_df.iloc[0:4].
Subsetting Data using Criteria
We can also select a subset of our data using criteria. For example, we can select all rows that have a year value of 2002:
Which produces the following output:
PYTHON
record_id month day year plot_id species_id sex hindfoot_length weight
33320 33321 1 12 2002 1 DM M 38 44
33321 33322 1 12 2002 1 DO M 37 58
33322 33323 1 12 2002 1 PB M 28 45
33323 33324 1 12 2002 1 AB NaN NaN NaN
33324 33325 1 12 2002 1 DO M 35 29
...
35544 35545 12 31 2002 15 AH NaN NaN NaN
35545 35546 12 31 2002 15 AH NaN NaN NaN
35546 35547 12 31 2002 10 RM F 15 14
35547 35548 12 31 2002 7 DO M 36 51
35548 35549 12 31 2002 5 NaN NaN NaN NaN
[2229 rows x 9 columns]Or we can select all rows that do not contain the year 2002:
We can define sets of criteria too:
Python Syntax Cheat Sheet
We can use the syntax below when querying data by criteria from a DataFrame. Experiment with selecting various subsets of the “surveys” data.
- Equals:
== - Not equals:
!= - Greater than:
> - Less than:
< - Greater than or equal to:
>= - Less than or equal to:
<=
Challenge - Queries
Select a subset of rows in the
surveys_dfDataFrame that contain data from the year 1999 and that contain weight values less than or equal to 8. How many rows did you end up with? What did your neighbor get?-
You can use the
isincommand in Python to query a DataFrame based upon a list of values as follows:Use the
isinfunction to find all plots that contain particular species in the “surveys” DataFrame. How many records contain these values? Experiment with other queries. Create a query that finds all rows with a weight value greater than or equal to 0.
The
~symbol in Python can be used to return the OPPOSITE of the selection that you specify. It is equivalent to is not in. Write a query that selects all rows with sex NOT equal to ‘M’ or ‘F’ in the “surveys” data.
-
OUTPUT
record_id month day year plot_id species_id sex hindfoot_length weight 29082 29083 1 16 1999 21 RM M 16.0 8.0 29196 29197 2 20 1999 18 RM M 18.0 8.0 29421 29422 3 15 1999 16 RM M 15.0 8.0 29903 29904 10 10 1999 4 PP M 20.0 7.0 29905 29906 10 10 1999 4 PP M 21.0 4.0If you are only interested in how many rows meet the criteria, the sum of
Truevalues could be used instead:OUTPUT
5 -
For example, using
PBandPL:OUTPUT
array([ 1, 10, 6, 24, 2, 23, 19, 12, 20, 22, 3, 9, 14, 13, 21, 7, 11, 15, 4, 16, 17, 8, 18, 5])OUTPUT
(24,) surveys_df[surveys_df["weight"] >= 0]-
OUTPUT
record_id month day year plot_id species_id sex hindfoot_length weight 13 14 7 16 1977 8 DM NaN NaN NaN 18 19 7 16 1977 4 PF NaN NaN NaN 33 34 7 17 1977 17 DM NaN NaN NaN 56 57 7 18 1977 22 DM NaN NaN NaN 76 77 8 19 1977 4 SS NaN NaN NaN ... ... ... ... ... ... ... ... ... ... 35527 35528 12 31 2002 13 US NaN NaN NaN 35543 35544 12 31 2002 15 US NaN NaN NaN 35544 35545 12 31 2002 15 AH NaN NaN NaN 35545 35546 12 31 2002 15 AH NaN NaN NaN 35548 35549 12 31 2002 5 NaN NaN NaN NaN [2511 rows x 9 columns]
When working through the solutions to the challenges above, you could introduce already that all these slice operations are actually based on a Boolean indexing operation (next section in the lesson). The filter provides for each record if it satisfies (True) or not (False). The slicing itself interprets the True/False of each record.
Using masks to identify a specific condition
A mask can be useful to locate where a particular
subset of values exist or don’t exist - for example, NaN, or “Not a
Number” values. To understand masks, we also need to understand
BOOLEAN objects in Python.
Boolean values include True or False. For
example,
When we ask Python whether x is greater than 5, it
returns False. This is Python’s way to say “No”. Indeed,
the value of x is 5, and 5 is not greater than 5.
To create a boolean mask:
- Set the True / False criteria
(e.g.
values > 5 = True) - Python will then assess each value in the object to determine whether the value meets the criteria (True) or not (False).
- Python creates an output object that is the same shape as the
original object, but with a
TrueorFalsevalue for each index location.
Let’s try this out. Let’s identify all locations in the survey data
that have null (missing or NaN) data values. We can use the
isnull method to do this. The isnull method
will compare each cell with a null value. If an element has a null
value, it will be assigned a value of True in the output
object.
A snippet of the output is below:
PYTHON
record_id month day year plot_id species_id sex hindfoot_length weight
0 False False False False False False False False True
1 False False False False False False False False True
2 False False False False False False False False True
3 False False False False False False False False True
4 False False False False False False False False True
[35549 rows x 9 columns]To select the rows where there are null values, we can use the mask as an index to subset our data as follows:
PYTHON
# To select just the rows with NaN values, we can use the 'any()' method
surveys_df[pd.isnull(surveys_df).any(axis=1)]Note that the weight column of our DataFrame contains
many null or NaN values. We will explore ways
of dealing with this in the next episode on Data Types and Formats.
We can run isnull on a particular column too. What does
the code below do?
PYTHON
# What does this do?
empty_weights = surveys_df[pd.isnull(surveys_df['weight'])]['weight']
print(empty_weights)Let’s take a minute to look at the statement above. We are using the
Boolean object pd.isnull(surveys_df['weight']) as an index
to surveys_df. We are asking Python to select rows that
have a NaN value of weight.
Challenge - Putting it all together
- Create a new DataFrame that only contains observations with sex values that are not female or male. Print the number of rows in this new DataFrame. Verify the result by comparing the number of rows in the new DataFrame with the number of rows in the surveys DataFrame where sex is null.
- Create a new DataFrame that contains only observations that are of sex male or female and where weight values are greater than 0. Create a stacked bar plot of average weight by plot with male vs female values stacked for each plot.
-
OUTPUT
2511OUTPUT
True PYTHON
# selection of the data with isin stack_selection = surveys_df[(surveys_df['sex'].isin(['M', 'F'])) & surveys_df["weight"] > 0.][["sex", "weight", "plot_id"]] # calculate the mean weight for each plot id and sex combination: stack_selection = stack_selection.groupby(["plot_id", "sex"]).mean().unstack() # and we can make a stacked bar plot from this: stack_selection.plot(kind='bar', stacked=True)
Referring to the challenge solution above, as we know the other values are all Nan values, we could also select all not null values:
PYTHON
stack_selection = surveys_df[(surveys_df['sex'].notnull()) &
surveys_df["weight"] > 0.][["sex", "weight", "plot_id"]]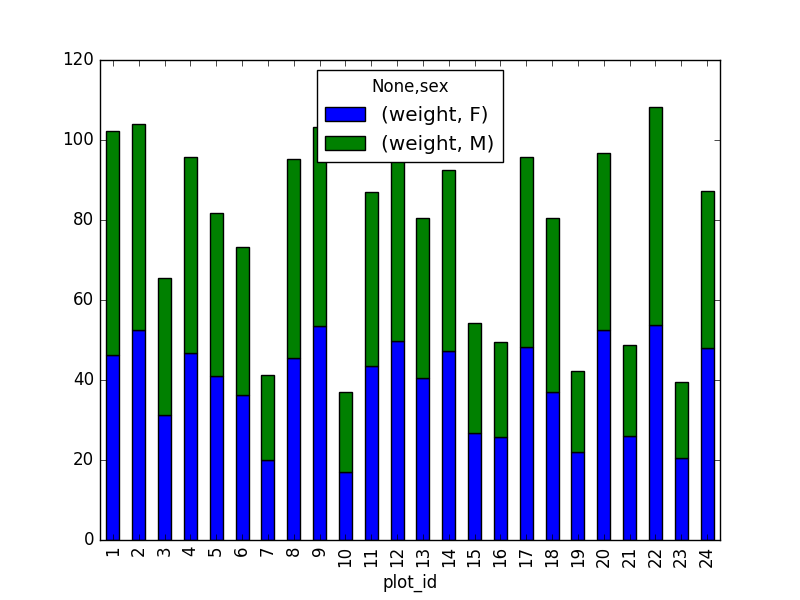
However, due to the unstack command, the legend header
contains two levels. In order to remove this, the column naming needs to
be simplified:
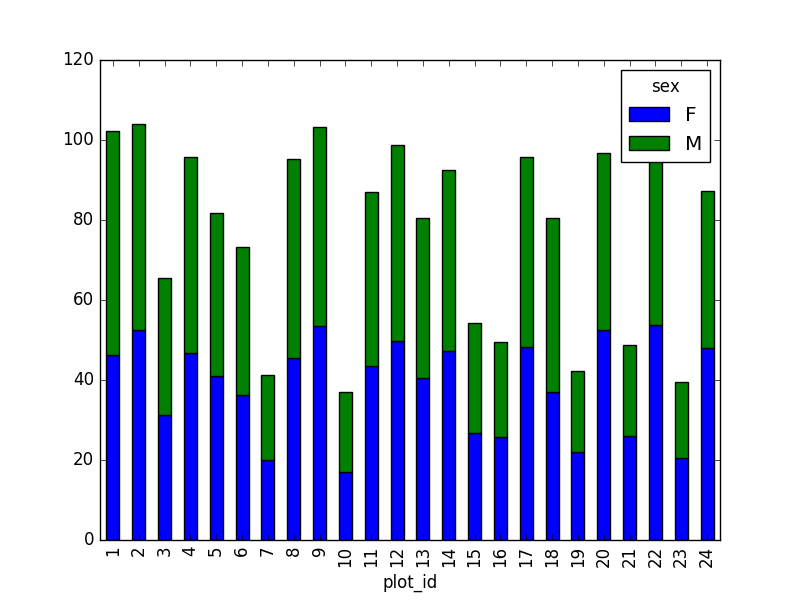
This is just a preview, more in next episode.
Key Points
- In Python, portions of data can be accessed using indices, slices, column headings, and condition-based subsetting.
- Python uses 0-based indexing, in which the first element in a list, tuple or any other data structure has an index of 0.
- Pandas enables common data exploration steps such as data indexing, slicing and conditional subsetting.
